Nanolive’s label-free, non-invasive imaging allows us access to the dynamics of biological processes, such as mitosis in living cells, with unprecedented resolution.
Here, we observe cell division in human mesenchymal stem cells using Nanolive’s 3D Cell Explorer. One image was captured every 15 secs for 2 h. The high-quality of the footage allows to identify and characterize the seven discrete steps of mitosis. In this blogpost, we will explain each phase individually, examine what is unique about the footage captured by Nanolive imaging, and discuss how these findings can help advance future studies.
What is mitosis and why is it important?
Mitosis is a form of eukaryotic cell division, in which one cell divides into two genetically identical daughter cells. It is a crucial part of the cell cycle, responsible for growth, wound healing, and the replacement of damaged cells.
Where does mitosis fit in the cell cycle?
A cell spends most of the duration of its life cycle in a stage known as interphase, which consists of three phases: Gap 1 (G1), Synthesis (S), and Gap 2 (G2; Fig. 1).
The G1 phase is dominated by cell growth; in this phase the cell is metabolically active, acquiring the nutrients required to build proteins and other molecules. Cells can sometimes leave G1 and enter a resting, or quiescent phase called G0, but, if the cell is actively dividing, it will proceed to enter the S phase, where the synthesis of DNA occurs and both chromosomes and centrosomes are duplicated.
After the S phase, the cell enters G2, where it adds volume to the cytoplasm and replicates many important organelles. It is possible to quantify changes in the cells dry mass content using Nanolive imaging, but that, is dealt with in another blogpost. Here, we will focus on describing the structures that are present at each discrete phase of mitosis (Fig. 2).
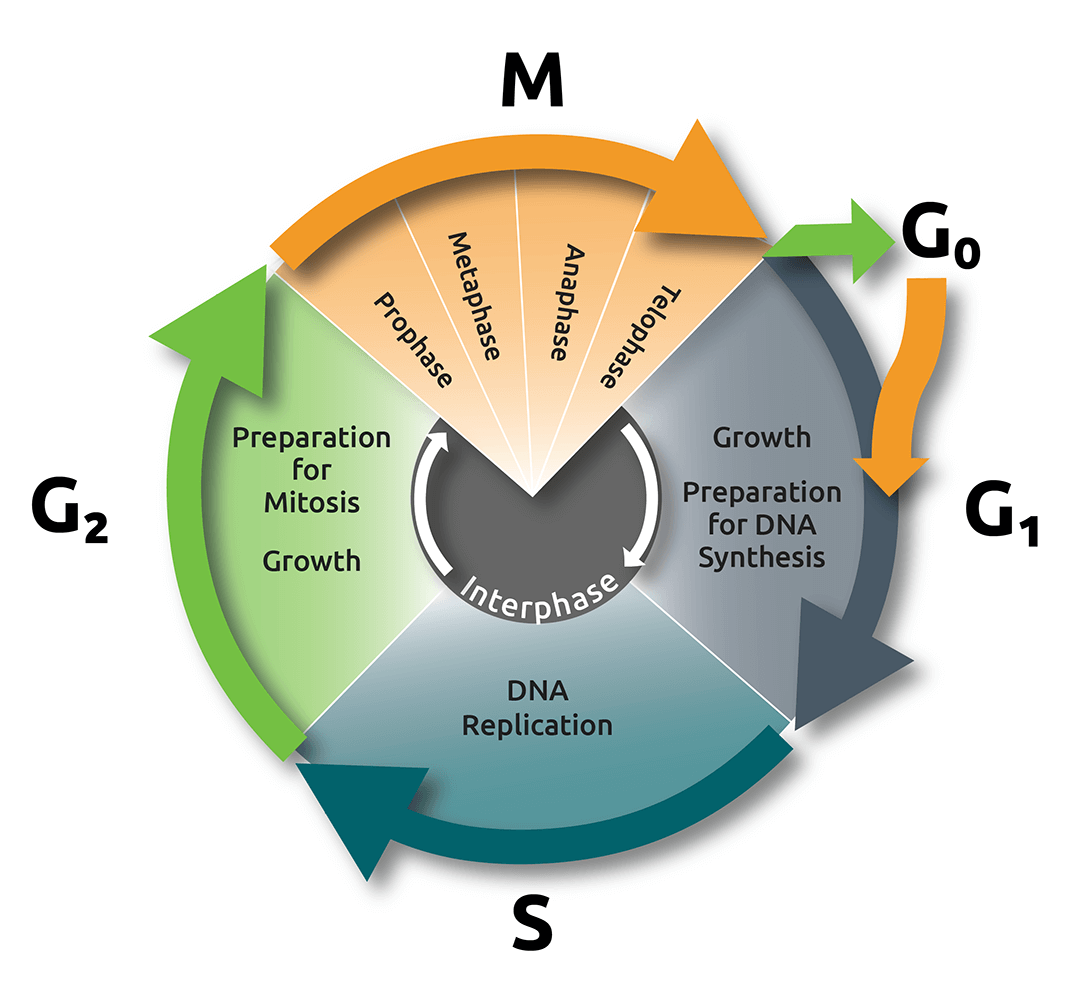
Fig.1.
The cell cycle and mitosis
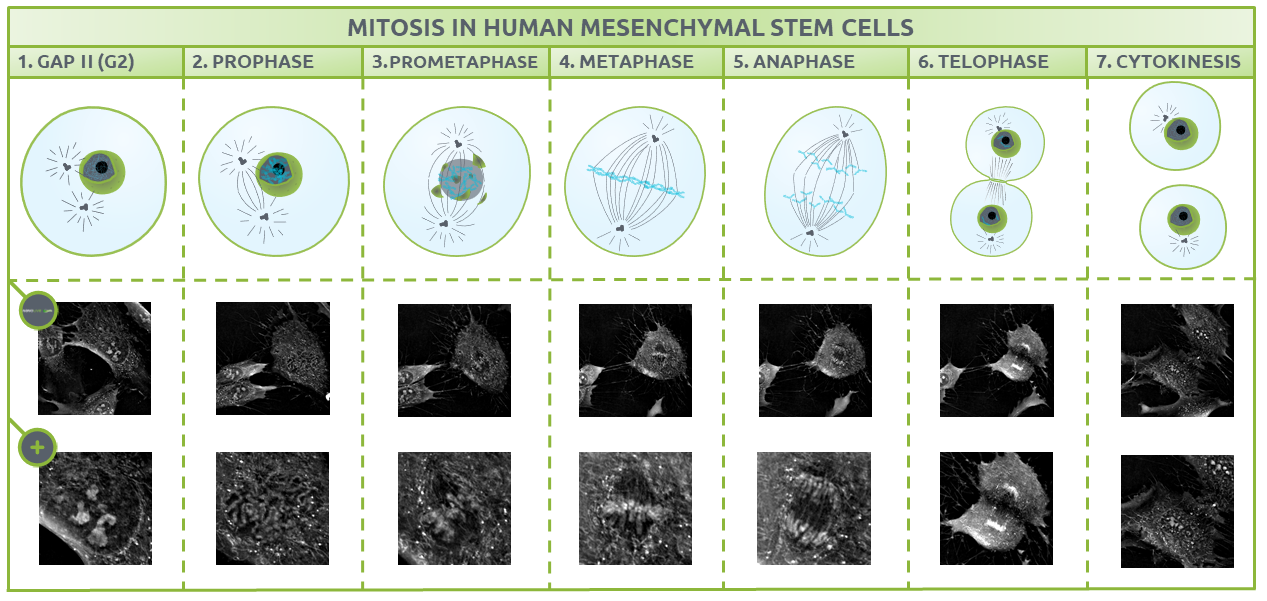
Fig.2.
The different phases of mitosis identified using Nanolive imaging
What is unique about the footage captured by Nanolive imaging?
Prophase
Once the cell is ready for division, it enters prophase, where nucleoli disappear and chromatin condenses into chromosomes, with two connected chromatids joined at the centromere (Fig. 3) (1,2). Chromosome formation from disorganised chromatin will ease the separation into different cells in further steps.
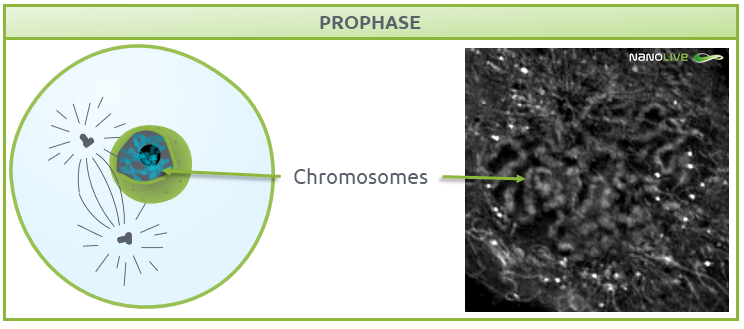
Fig.3.
Characteristic structures present during prophase
Prometaphase
During prometaphase, the nuclear membrane breaks down into small-sized membrane vesicles (Fig. 4) to create space for microtubules to attach to protein structures called kinetochores (disc-shaped structures associated with duplicated chromatids), which help centrosomes pull the chromosomes apart (3-5). Mitotic spindles work in conjunction with kinetochores, centrosomes, and astral microtubules, the latter of which radiate from the centrosome towards the cell edge. All these structures work as a coordinated machinery, and cell organelles like mitochondria are usually absent in the area they delimit (6,7).
![]() Note the disappearance of nuclear membrane, and the absence of mitochondria where we expect the mitotic spindles during this phase.
Note the disappearance of nuclear membrane, and the absence of mitochondria where we expect the mitotic spindles during this phase.
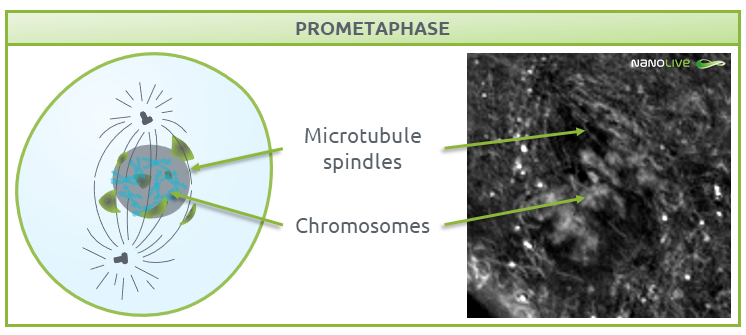
Fig.4.
Characteristic structures present during prometaphase
Metaphase
In metaphase, chromosomes line up along the metaphase plate; a plane created by the equal tension that is applied by the two sister chromatids that are attached to each chromosome and pulled by the microtubules attached to the two spindle poles (Fig. 5).
![]() Note the increase in refractive index at the metaphase plate. This reflects the high number of proteins in the microtubules of the mitotic spindle and the kinetochores.
Note the increase in refractive index at the metaphase plate. This reflects the high number of proteins in the microtubules of the mitotic spindle and the kinetochores.
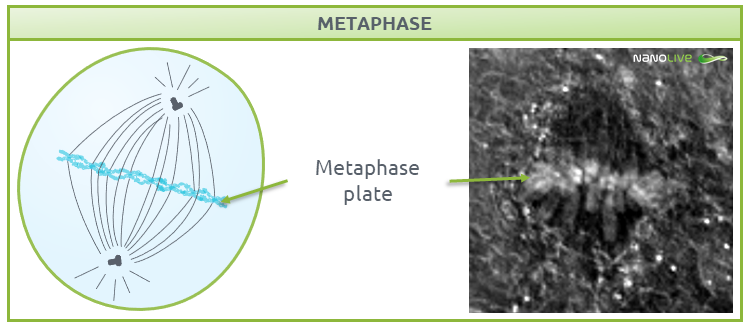
Fig.5.
Characteristic structures present during metaphase.
Up until now, the steps must occur in order, but after metaphase, the remaining steps (anaphase, telophase, and cytokinesis) can overlap or occur simultaneously (8).
Anaphase
During anaphase, chromosomes break at the centromeres and sister chromatids migrate to opposite ends of the cell (Fig. 6). The non-kinetochore spindle fibres (the microtubules that are not attached to the chromosomes), provide the tension needed for chromatid migration and work together with motor proteins to help chromosomes move along microtubule tracks.
![]() Note the position of the sister chromatids.
Note the position of the sister chromatids.
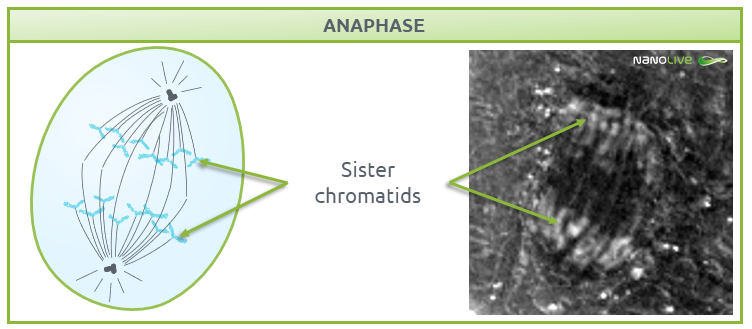
Fig.6.
Characteristic structures present during anaphase.
Telophase
During telophase (Fig. 7), the nuclear membrane is reformed to protect the DNA. As the cell needs new gene products, the DNA must be accessible. DNA replication requires decondensed chromatin, so chromosomes de-compact into chromatin, and the nucleoli containing ribosomes reappear.
![]() Note the presence of the contractile ring.
Note the presence of the contractile ring.
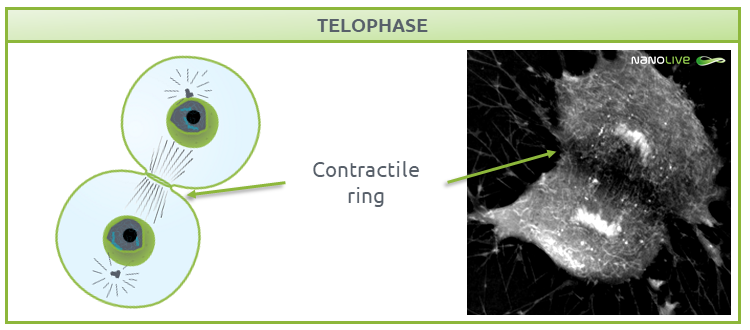
Fig.7.
Characteristic structures present during telophase.
Cytokinesis
Cytokinesis is the final step of the mitotic process, where the two daughter cells are separated (Fig. 8). Each of these daughter cells contain a complete set of chromosomes and are ready to enter G1 phase.
During this process, a complex structure called the contractile ring separates the two fully functional daughter cells, ending the cycle (9). Read more about cytokinesis in mesenchymal stem cells in our dedicated post here.
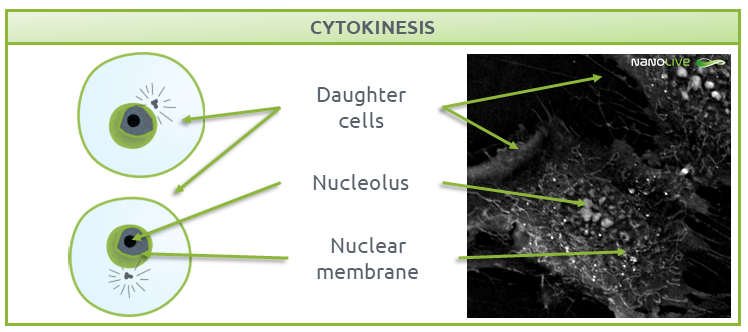
Fig.8.
Characteristic structures present during Cytokinesis.
What do these findings offer researchers in the future?
This technique should be of interest to stem cell researchers who study regenerative processes, where the aim is to switch dormant cells back to replicative cells (10, 11). The ability to identify the phase of mitosis that the cells are in with high confidence, could also help researchers understand how DNA damage and repair mechanisms function in stem cells (12).
References
(1) Shishova, K. V. et al. Acta Naturae 3, 100-106 (2011).
(2) Cooper, G. M. The nucleus during mitosis in the Cell: A Molecular Approach. Sinauer Associates, Sunderland, MA, USA, ed. 2. (2000).
(3) Maiato, H. et al. J. Cell Sci. 117, 5461-5477 (2004).
(4) Heald, R. Cell. 102, 399-402 (2000).
(5) Mcintosh, J. R. & Pfarr, C. M. J. Cell Biol. 115, 577-585 (1991).
(6) Ouellet, J. & Barral, Y. J. Cell Biol. 196, 305-313 (2012).
(7) Robbins, E. & Gonatas, N. K. J. Cell Biol. 21, 429-463 (1964).
(8) Mishima, M. et al. Nature. 430, 908-913 (2004).
(9) Schroeder, T.E. Z Zellforsch Mikrosk Anat.109 (4) 431-449 (1970).
(10) Pittenger, M. F. et al. NPJ Regen. Med. 4, 1-15 (2019).
(11) Chen, Y. et al. Int. J. Biochem. Cell Biol. 40, 815-820 (2008).
(12) Beerman, I. et al. Cell Stem Cell 15, 37-50 (2014).
Read our latest news
Cytotoxic Drug Development Application Note
Discover how Nanolive’s LIVE Cytotoxicity Assay transforms cytotoxic drug development through high-resolution, label-free quantification of cell health and death. Our application note explores how this advanced technology enables real-time monitoring of cell death...
Investigative Toxicology Application Note
Our groundbreaking approach offers a label-free, high-content imaging solution that transforms the way cellular health, death, and phenotypic responses are monitored and quantified. Unlike traditional cytotoxicity assays, Nanolive’s technology bypasses the limitations...
Phenotypic Cell Health and Stress Application Note
Discover the advanced capabilities of Nanolive’s LIVE Cytotoxicity Assay in an application note. This document presents a detailed exploration of how our innovative, label-free technology enables researchers to monitor phenotypic changes and detect cell stress...



