Scientific Posters
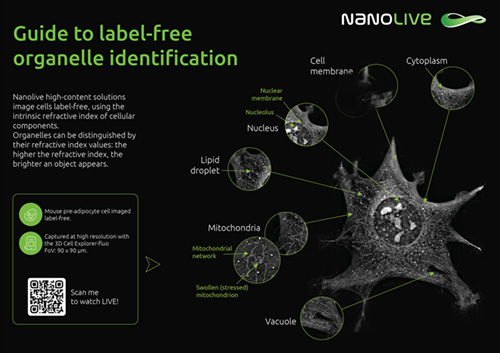
Guide to label-free organelle identification
Nanolive platforms image cells label-free, using the intrinsic refractive index of cellular components. The higher the refractive index, the brighter an object appears, which allows us to identify many different organelles without fluorescence or dyes.
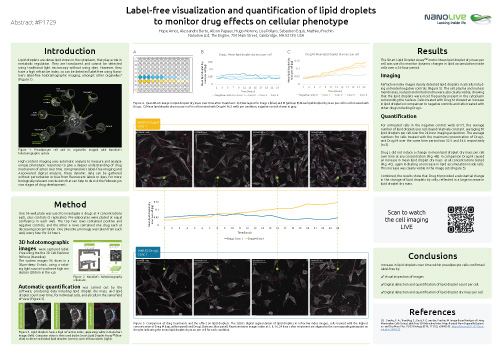
Label-free lipid droplet quantification
Lipid droplets are dense lipid stores in the cytoplasm, that play a role in metabolic regulation. They are translucent and cannot be detected using traditional light microscopy without using dyes. However, they have a high refractive index, so can be detected label-free using Nanolive imaging, confirmed by correlative fluorescence imaging. The Smart Lipid Droplet Assay LIVE (SDLA) segments lipid droplets and cell boundaries automatically to quantify changes in lipid droplets over time.
Past Publications
Quantifying dry mass dynamics in lipid droplets using Nanolive cell imaging
Lipid droplets (LDs) play a key role in energy and membrane lipid metabolism, yet very little is known about the dynamic processes involved in their formation. Capturing the appearance and early growth of LDs has so far been elusive, largely due to method limitations. Nanolive cell imaging is label-free, and so it circumvents the temporal restrictions that phototoxicity imposes during classical fluorescence microscopy, making it possible to perform high-frequency imaging over long periods of time, without perturbing organelle biology. Here, we investigate LD dry mass dynamics.
Click below to download all past posters.

Inducing and quantifying the accumulation of late endosomes in a Niemann-Pick Type C cell phenotype
The endocytic pathway internalizes molecules from the plasma membrane into distinct membrane-bound compartments, which can be recycled, disseminated or degraded. Disruptions in these trafficking pathways can cause serious diseases. One such example of this is Niemann-Pick Type C disease, a rare progressive genetic disorder characterized by an abnormal accumulation of cholesterol and other fatty substances inside of cells. Here, we use the compound U18666A to simulate the Niemann-Pick Type C cell phenotype and use Nanolive label-free cell imaging to quantify the accumulation of late endosomes.
Click below to download all past posters.

Investigating fine scale dry mass dynamics during mitosis using Nanolive cell imaging
Mitosis has been extensively studied using a range of imaging techniques, yet key pieces of information remain elusive. One topic that remains controversial, is how cell volume changes during mitosis. Some studies show that cell volume diminishes, while others demonstrate that cells swell during mitosis.
Nanolive cell imaging is a label-free imaging technique that produces high quality refractive index (RI) maps in 3D. It is therefore well suited to study dynamic biological processes in fine detail. In this study, we quantify cellular dry mass dynamics before, during, and after mitosis.
Click below to download all past posters.

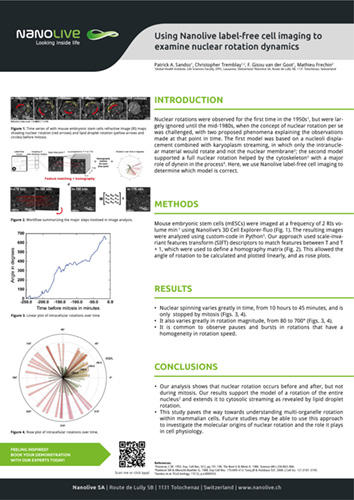
Using Nanolive label-free cell imaging to examine nuclear rotation dynamics
Nuclear rotations were observed for the first time in the 1950s, but were largely ignored until the mid-1980s, when the concept of nuclear rotation per se was challenged, with two proposed phenomena explaining the observations made at that point in time. The first model was based on a nucleoli displacement combined with karyoplasm streaming, in which only the intranuclear material would rotate and not the nuclear membrane; the second model supported a full nuclear rotation helped by the cytoskeleton with a major role of dynein in the process. Here, we use Nanolive label-free cell imaging to determine which model is correct.
Click below to download all past posters.
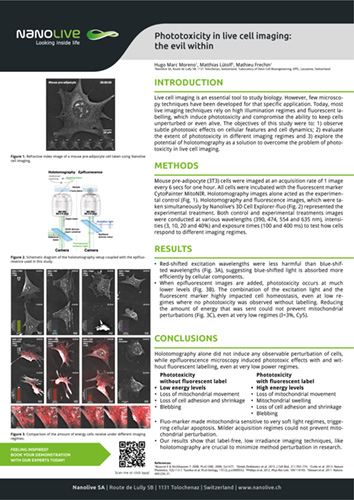
Phototoxicity in live cell imaging: the evil within
Live cell imaging is an essential tool to study biology. However, few microscopy techniques have been developed for that specific application. Today, most live imaging techniques rely on high illumination regimes and fluorescent labelling, which induce phototoxicity and compromise the ability to keep cells unperturbed or even alive. The objectives of this study were to: 1) observe subtle phototoxic effects on cellular features and cell dynamics; 2) evaluate the extent of phototoxicity in different imaging regimes and 3) explore the potential of holotomography as a solution to overcome the problem of phototoxicity in live cell imaging.
Click below to download all past posters.